6.21 Text Processing Model
A text processing model uses ctx.settings arguments to specify Oracle Text attribute settings.
Example 6-26 Building a Text Processing Model
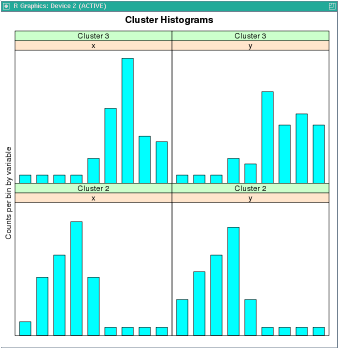
This example builds an ore.odmKMeans model that processes text. It uses the odm.settings and ctx.settings arguments. The figure following the example shows the output of the histogram(km.mod1) function.
x <- rbind(matrix(rnorm(100, sd = 0.3), ncol = 2),
matrix(rnorm(100, mean = 1, sd = 0.3), ncol = 2))
colnames(x) <- c("x", "y")
X <- ore.push (data.frame(x))
km.mod1 <- NULL
km.mod1 <- ore.odmKMeans(~., X, num.centers = 2)
km.mod1
summary(km.mod1)
rules(km.mod1)
clusterhists(km.mod1)
histogram(km.mod1)
km.res1 <- predict(km.mod1,X,type="class",supplemental.cols=c("x","y"))
head(km.res1,3)
km.res1.local <- ore.pull(km.res1)
plot(data.frame(x = km.res1.local$x,
y = km.res1.local$y),
col = km.res1.local$CLUSTER_ID)
points(km.mod1$centers2, col = rownames(km.mod1$centers2), pch = 8, cex=2)
head(predict(km.mod1,X))
head(predict(km.mod1,X,type=c("class","raw"),supplemental.cols=c("x","y")),3)
head(predict(km.mod1,X,type="raw",supplemental.cols=c("x","y")),3)
# Text processing with ore.odmKMeans.
title <- c('Aids in Africa: Planning for a long war',
'Mars rover maneuvers for rim shot',
'Mars express confirms presence of water at Mars south pole',
'NASA announces major Mars rover finding',
'Drug access, Asia threat in focus at AIDS summit',
'NASA Mars Odyssey THEMIS image: typical crater',
'Road blocks for Aids')
response <- c('Aids', 'Mars', 'Mars', 'Mars', 'Aids', 'Mars', 'Aids')
# Text contents in a character column.
KM_TEXT <- ore.push(data.frame(CUST_ID = seq(length(title)),
RESPONSE = response, TITLE = title))
# Create a text policy (CTXSYS.CTX_DDL privilege is required).
ore.exec("Begin ctx_ddl.create_policy('ESA_TXTPOL'); End;")
# Specify POLICY_NAME, MIN_DOCUMENTS, MAX_FEATURES and
# text column attributes.
km.mod <- ore.odmKMeans( ~ TITLE, data = KM_TEXT, num.centers = 2L,
odm.settings = list(ODMS_TEXT_POLICY_NAME = "ESA_TXTPOL",
ODMS_TEXT_MIN_DOCUMENTS = 1,
ODMS_TEXT_MAX_FEATURES = 3,
kmns_distance = "dbms_data_mining.kmns_cosine",
kmns_details = "kmns_details_all"),
ctx.settings = list(TITLE = "TEXT(TOKEN_TYPE:STEM)"))
summary(km.mod)
settings(km.mod)
print(predict(km.mod, KM_TEXT, supplemental.cols = "RESPONSE"), digits = 3L)
ore.exec("Begin ctx_ddl.drop_policy('ESA_TXTPOL'); End;")Listing for This Example
R> x <- rbind(matrix(rnorm(100, sd = 0.3), ncol = 2),
+ matrix(rnorm(100, mean = 1, sd = 0.3), ncol = 2))
R> colnames(x) <- c("x", "y")
R>
R> X <- ore.push (data.frame(x))
R> km.mod1 <- NULL
R> km.mod1 <- ore.odmKMeans(~., X, num.centers = 2)
R> km.mod1
Call:
ore.odmKMeans(formula = ~., data = X, num.centers = 2)
Settings:
value
clus.num.clusters 2
block.growth 2
conv.tolerance 0.01
details details.all
distance euclidean
iterations 3
min.pct.attr.support 0.1
num.bins 10
random.seed 0
split.criterion variance
odms.missing.value.treatment odms.missing.value.auto
odms.sampling odms.sampling.disable
prep.auto ON
R> summary(km.mod1)
Call:
ore.odmKMeans(formula = ~., data = X, num.centers = 2)
Settings:
value
clus.num.clusters 2
block.growth 2
conv.tolerance 0.01
details details.all
distance euclidean
iterations 3
min.pct.attr.support 0.1
num.bins 10
random.seed 0
split.criterion variance
odms.missing.value.treatment odms.missing.value.auto
odms.sampling odms.sampling.disable
prep.auto ON
Centers:
x y
2 -0.07638266 0.04449368
3 0.98493306 1.00864399
R> rules(km.mod1)
cluster.id rhs.support rhs.conf lhr.support lhs.conf lhs.var lhs.var.support lhs.var.conf predicate
1 1 100 1.0 92 0.86 x 86 0.2222222 x <= 1.2209
2 1 100 1.0 92 0.86 x 86 0.2222222 x >= -.6188
3 1 100 1.0 86 0.86 y 86 0.4444444 y <= 1.1653
4 1 100 1.0 86 0.86 y 86 0.4444444 y > -.3053
5 2 50 0.5 48 0.96 x 48 0.0870793 x <= .4324
6 2 50 0.5 48 0.96 x 48 0.0870793 x >= -.6188
7 2 50 0.5 48 0.96 y 48 0.0893300 y <= .5771
8 2 50 0.5 48 0.96 y 48 0.0893300 y > -.5995
9 3 50 0.5 49 0.98 x 49 0.0852841 x <= 1.7465
10 3 50 0.5 49 0.98 x 49 0.0852841 x > .4324
11 3 50 0.5 50 0.98 y 49 0.0838225 y <= 1.7536
12 3 50 0.5 50 0.98 y 49 0.0838225 y > .2829
R> clusterhists(km.mod1)
cluster.id variable bin.id lower.bound upper.bound label count
1 1 x 1 -0.61884662 -0.35602715 -.6188466:-.3560272 6
2 1 x 2 -0.35602715 -0.09320769 -.3560272:-.0932077 17
3 1 x 3 -0.09320769 0.16961178 -.0932077:.1696118 15
4 1 x 4 0.16961178 0.43243125 .1696118:.4324312 11
5 1 x 5 0.43243125 0.69525071 .4324312:.6952507 8
6 1 x 6 0.69525071 0.95807018 .6952507:.9580702 17
7 1 x 7 0.95807018 1.22088965 .9580702:1.2208896 18
8 1 x 8 1.22088965 1.48370911 1.2208896:1.4837091 4
9 1 x 9 1.48370911 1.74652858 1.4837091:1.7465286 4
10 1 y 1 -0.89359597 -0.59946141 -.893596:-.5994614 2
11 1 y 2 -0.59946141 -0.30532685 -.5994614:-.3053269 4
12 1 y 3 -0.30532685 -0.01119230 -.3053269:-.0111923 11
13 1 y 4 -0.01119230 0.28294226 -.0111923:.2829423 24
14 1 y 5 0.28294226 0.57707682 .2829423:.5770768 13
15 1 y 6 0.57707682 0.87121138 .5770768:.8712114 12
16 1 y 7 0.87121138 1.16534593 .8712114:1.1653459 26
17 1 y 8 1.16534593 1.45948049 1.1653459:1.4594805 5
18 1 y 9 1.45948049 1.75361505 1.4594805:1.753615 3
19 2 x 1 -0.61884662 -0.35602715 -.6188466:-.3560272 6
20 2 x 2 -0.35602715 -0.09320769 -.3560272:-.0932077 17
21 2 x 3 -0.09320769 0.16961178 -.0932077:.1696118 15
22 2 x 4 0.16961178 0.43243125 .1696118:.4324312 10
23 2 x 5 0.43243125 0.69525071 .4324312:.6952507 2
24 2 x 6 0.69525071 0.95807018 .6952507:.9580702 0
25 2 x 7 0.95807018 1.22088965 .9580702:1.2208896 0
26 2 x 8 1.22088965 1.48370911 1.2208896:1.4837091 0
27 2 x 9 1.48370911 1.74652858 1.4837091:1.7465286 0
28 2 y 1 -0.89359597 -0.59946141 -.893596:-.5994614 2
29 2 y 2 -0.59946141 -0.30532685 -.5994614:-.3053269 4
30 2 y 3 -0.30532685 -0.01119230 -.3053269:-.0111923 11
31 2 y 4 -0.01119230 0.28294226 -.0111923:.2829423 24
32 2 y 5 0.28294226 0.57707682 .2829423:.5770768 9
33 2 y 6 0.57707682 0.87121138 .5770768:.8712114 0
34 2 y 7 0.87121138 1.16534593 .8712114:1.1653459 0
35 2 y 8 1.16534593 1.45948049 1.1653459:1.4594805 0
36 2 y 9 1.45948049 1.75361505 1.4594805:1.753615 0
37 3 x 1 -0.61884662 -0.35602715 -.6188466:-.3560272 0
38 3 x 2 -0.35602715 -0.09320769 -.3560272:-.0932077 0
39 3 x 3 -0.09320769 0.16961178 -.0932077:.1696118 0
40 3 x 4 0.16961178 0.43243125 .1696118:.4324312 1
41 3 x 5 0.43243125 0.69525071 .4324312:.6952507 6
42 3 x 6 0.69525071 0.95807018 .6952507:.9580702 17
43 3 x 7 0.95807018 1.22088965 .9580702:1.2208896 18
44 3 x 8 1.22088965 1.48370911 1.2208896:1.4837091 4
45 3 x 9 1.48370911 1.74652858 1.4837091:1.7465286 4
46 3 y 1 -0.89359597 -0.59946141 -.893596:-.5994614 0
47 3 y 2 -0.59946141 -0.30532685 -.5994614:-.3053269 0
48 3 y 3 -0.30532685 -0.01119230 -.3053269:-.0111923 0
49 3 y 4 -0.01119230 0.28294226 -.0111923:.2829423 0
50 3 y 5 0.28294226 0.57707682 .2829423:.5770768 4
51 3 y 6 0.57707682 0.87121138 .5770768:.8712114 12
52 3 y 7 0.87121138 1.16534593 .8712114:1.1653459 26
53 3 y 8 1.16534593 1.45948049 1.1653459:1.4594805 5
54 3 y 9 1.45948049 1.75361505 1.4594805:1.753615 3
R> histogram(km.mod1)
R>
R> km.res1 <- predict(km.mod1, X, type="class", supplemental.cols = c("x","y"))
R> head(km.res1, 3)
x y CLUSTER_ID
1 -0.43646407 0.26201831 2
2 -0.02797831 0.07319952 2
3 0.11998373 -0.08638716 2
R> km.res1.local <- ore.pull(km.res1)
R> plot(data.frame(x = km.res1.local$x,
+ y = km.res1.local$y),
+ col = km.res1.local$CLUSTER_ID)
R> points(km.mod1$centers2, col = rownames(km.mod1$centers2), pch = 8, cex = 2)
R>
R> head(predict(km.mod1, X))
'2' '3' CLUSTER_ID
1 0.9992236 0.0007763706 2
2 0.9971310 0.0028690375 2
3 0.9974216 0.0025783939 2
4 0.9997335 0.0002665114 2
5 0.9917773 0.0082226599 2
6 0.9771667 0.0228333398 2
R> head(predict(km.mod1,X,type=c("class","raw"),supplemental.cols=c("x","y")),3)
'2' '3' x y CLUSTER_ID
1 0.9992236 0.0007763706 -0.43646407 0.26201831 2
2 0.9971310 0.0028690375 -0.02797831 0.07319952 2
3 0.9974216 0.0025783939 0.11998373 -0.08638716 2
R> head(predict(km.mod1,X,type="raw",supplemental.cols=c("x","y")),3)
x y '2' '3'
1 -0.43646407 0.26201831 0.9992236 0.0007763706
2 -0.02797831 0.07319952 0.9971310 0.0028690375
3 0.11998373 -0.08638716 0.9974216 0.0025783939R>
R>
R> # Text processing with ore.odmKMeans.
R> title <- c('Aids in Africa: Planning for a long war',
+ 'Mars rover maneuvers for rim shot',
+ 'Mars express confirms presence of water at Mars south pole',
+ 'NASA announces major Mars rover finding',
+ 'Drug access, Asia threat in focus at AIDS summit',
+ 'NASA Mars Odyssey THEMIS image: typical crater',
+ 'Road blocks for Aids')
R> response <- c('Aids', 'Mars', 'Mars', 'Mars', 'Aids', 'Mars', 'Aids')
R>
R> # Text contents in a character column.
R> KM_TEXT <- ore.push(data.frame(CUST_ID = seq(length(title)),
+ RESPONSE = response, TITLE = title))
R>
R> # Create a text policy (CTXSYS.CTX_DDL privilege is required).
R> ore.exec("Begin ctx_ddl.create_policy('ESA_TXTPOL'); End;")
R>
R> # Specify POLICY_NAME, MIN_DOCUMENTS, MAX_FEATURES and
R> # text column attributes.
R> km.mod <- ore.odmKMeans( ~ TITLE, data = KM_TEXT, num.centers = 2L,
+ odm.settings = list(ODMS_TEXT_POLICY_NAME = "ESA_TXTPOL",
+ ODMS_TEXT_MIN_DOCUMENTS = 1,
+ ODMS_TEXT_MAX_FEATURES = 3,
+ kmns_distance = "dbms_data_mining.kmns_cosine",
+ kmns_details = "kmns_details_all"),
+ ctx.settings = list(TITLE="TEXT(TOKEN_TYPE:STEM)"))
R> summary(km.mod)
Call:
ore.odmKMeans(formula = ~TITLE, data = KM_TEXT, num.centers = 2L,
odm.settings = list(ODMS_TEXT_POLICY_NAME = "ESA_TXTPOL",
ODMS_TEXT_MIN_DOCUMENTS = 1, ODMS_TEXT_MAX_FEATURES = 3,
kmns_distance = "dbms_data_mining.kmns_cosine",
kmns_details = "kmns_details_all"),
ctx.settings = list(TITLE = "TEXT(TOKEN_TYPE:STEM)"))
Settings:
value
clus.num.clusters 2
block.growth 2
conv.tolerance 0.01
details details.all
distance cosine
iterations 3
min.pct.attr.support 0.1
num.bins 10
random.seed 0
split.criterion variance
odms.missing.value.treatment odms.missing.value.auto
odms.sampling odms.sampling.disable
odms.text.max.features 3
odms.text.min.documents 1
odms.text.policy.name ESA_TXTPOL
prep.auto ON
Centers:
TITLE.MARS TITLE.NASA TITLE.ROVER TITLE.AIDS
2 0.5292307 0.7936566 0.7936566 NA
3 NA NA NA 1
R> settings(km.mod)
SETTING_NAME SETTING_VALUE SETTING_TYPE
1 ALGO_NAME ALGO_KMEANS INPUT
2 CLUS_NUM_CLUSTERS 2 INPUT
3 KMNS_BLOCK_GROWTH 2 INPUT
4 KMNS_CONV_TOLERANCE 0.01 INPUT
5 KMNS_DETAILS KMNS_DETAILS_ALL INPUT
6 KMNS_DISTANCE KMNS_COSINE INPUT
7 KMNS_ITERATIONS 3 INPUT
8 KMNS_MIN_PCT_ATTR_SUPPORT 0.1 INPUT
9 KMNS_NUM_BINS 10 INPUT
10 KMNS_RANDOM_SEED 0 DEFAULT
11 KMNS_SPLIT_CRITERION KMNS_VARIANCE INPUT
12 ODMS_MISSING_VALUE_TREATMENT ODMS_MISSING_VALUE_AUTO DEFAULT
13 ODMS_SAMPLING ODMS_SAMPLING_DISABLE DEFAULT
14 ODMS_TEXT_MAX_FEATURES 3 INPUT
15 ODMS_TEXT_MIN_DOCUMENTS 1 INPUT
16 ODMS_TEXT_POLICY_NAME ESA_TXTPOL INPUT
17 PREP_AUTO ON INPUT
R> print(predict(km.mod, KM_TEXT, supplemental.cols = "RESPONSE"), digits = 3L)
'2' '3' RESPONSE CLUSTER_ID
1 0.0213 0.9787 Aids 3
2 0.9463 0.0537 Mars 2
3 0.9325 0.0675 Mars 2
4 0.9691 0.0309 Mars 2
5 0.0213 0.9787 Aids 3
6 0.9463 0.0537 Mars 2
7 0.0213 0.9787 Aids 3
R>
R> ore.exec("Begin ctx_ddl.drop_policy('ESA_TXTPOL'); End;")